Nipah is one of the deadliest viruses in the world and considered one of the top future pandemic risks.
We're hosting a protein design competition on Proteinbase where people can design binders against the virus and we will test 1200 of those designs in our lab to see if they actually work.
Last year's EGFR competition had a 13.5% hit rate overall (54 out of 400 proteins) -- what will it be for Nipah virus? And which model will have the highest hit rate?

The response was incredible: Over 10’000 designs were submitted, over 5 times the number of designs in the past 2 competitions combined, with more than 650 total participants.
We wrote a blog post with analysis around which protein design models were used, what types of proteins designed etc here: https://www.adaptyvbio.com/blog/nipah-submissions/
While the protein designs are being tested in our lab, you can now predict which protein design model will yield the highest hit rates! Check out all the submissions here and then place your bets: https://proteinbase.com/competitions/adaptyv-nipah-competition
Some examples:
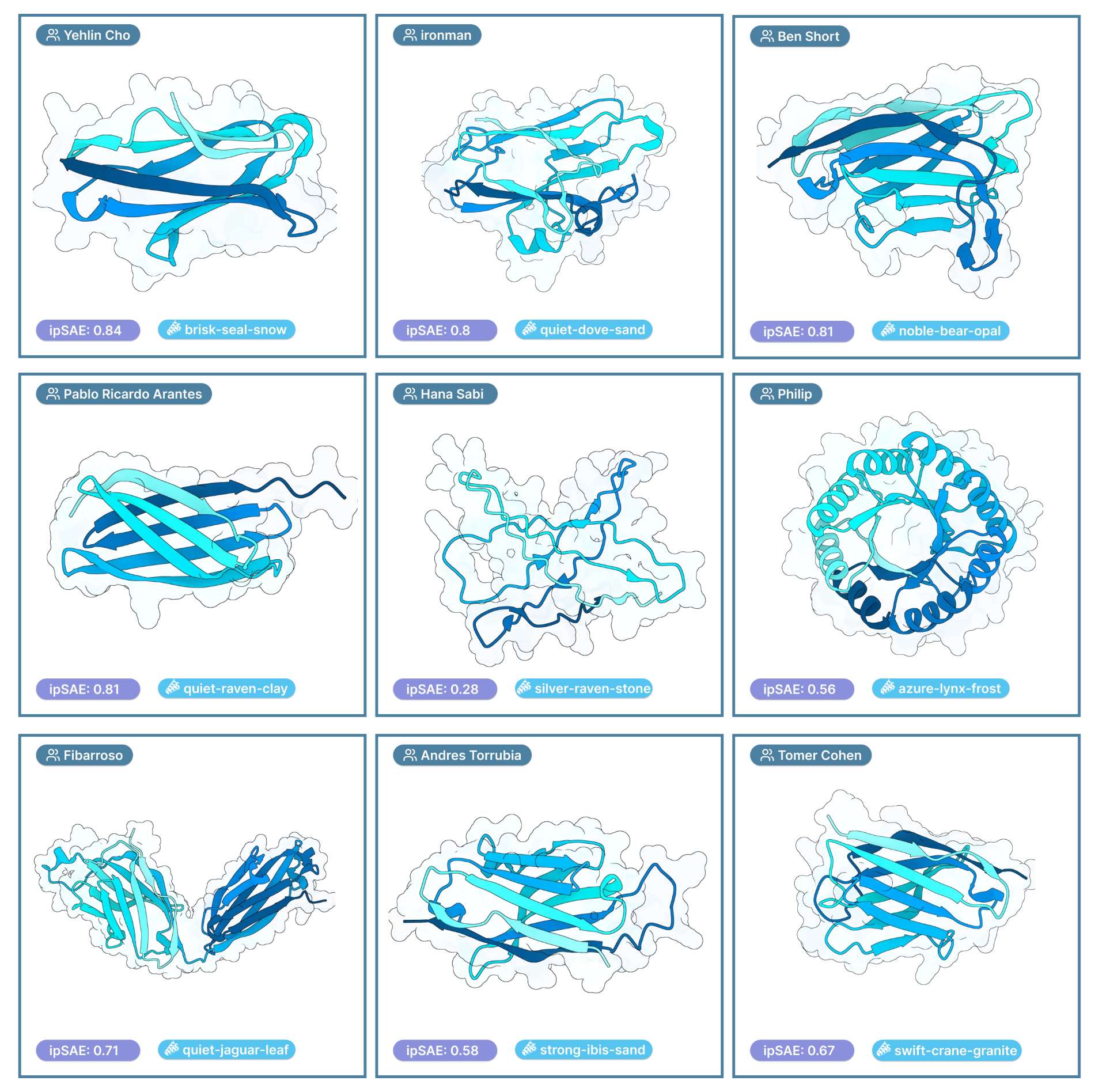
How will this be resolved?
We are publishing the lab results on Jan 18 2026. Binders are validated using SPR (Surface Plasmon Resonance) in our lab at Adaptyv Bio. All results will be open-sourced on Proteinbase.
Why don't I find protein design model X, Y or Z?
You can just add any protein design model from the Nipah binder competition as an answer to the question.